Patrik R. Callis
Seeking the Secret of Enzymes
Enzymes enormously accelerate the rates of chemical reactions over the rates of the same reactions in water (by 108-1015-fold), but the precise manner by which enzymes accomplish this in detail is still considered an open question.
We are currently making a seamless transition from obtaining a detailed understanding of how the intense internal electric fields in proteins profoundly affect the properties of tryptophan fluorescence, towards a better understanding and more detailed view of how enzymes attain their astronomical acceleration of biochemical reactions.
This includes performing classical and quantum molecular dynamics computations on the active sites of many enzymes, with the goal of observing unbiased enzymatic reaction events.
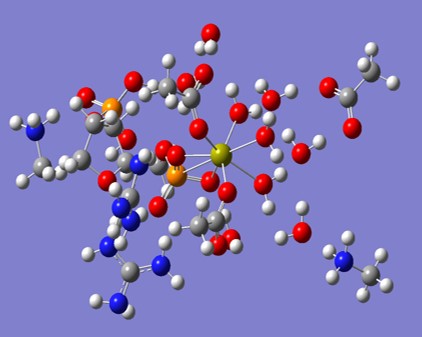
The image below shows a typical cluster of molecules from the active site of Staph. Nuclease, upon which we have performed quantum molecular dynamics computations.
Videos of Quantum Dyamics
Under Construction
Preprint:
Water as a reactant in the first step of triosephosphate isomerase catalysis
MovieS1 Movie of Hydroxide abstracting proR H
Caption: Movie S1 (separate file to be uploaded separately). File: MovieS1-DHAP-OH.mp4 Caption: Movie of ADMP trajectory of a hydroxide ion abstracting the pro-R proton (atom 22) from DHAP in the presence of Glu165. Route card = # b3lyp/6-31g(d,p) admp=(maxpoints=2000,nke=37052)
MovieS2 Optimization of Water Dimer in High Field
Caption: MP4 movie of Gaussian 09 energy minimization computation of a gas phase water dimer in an applied constant electric field of 0.065 au. Route = # opt b3lyp/cc-pVTZ nosym field=x-650 scf=(xqc,maxconventionalcycles=512
Recent Publications
-
Muiño, P. L., Callis, P.R., (2020) Water as an Essential Cofactor for All Enzymes, Biophys. J. v. 118 i.3 p.535A-535A
- Purnell, G.E., McNally M.T., Callis, P. R., Walker, R.A., (2020) Buried Liquid Interfaces as a Form of Chemistry in Confinement: The Case of 4-Dimethylaminobenzonitrile at the Silica-Aqueous Interface. J. Am. Chem. Soc. v. 142 i. 5 p. 2375-2385
-
MIKHAYLOV, A., DE REGUARDATI, S., PAHAPILL, J., Callis, P. R., Kohler, B., REBANE, A. (2018) Two-photon absorption spectra of fluorescent isomorphic DNA base analogs. Biomedical Otics Express; Optical Society of America: v. 9 i. 2 p. 6
- Uudsemaa, M., Trummal, A., de Reguardati, S., Callis, P., Rebane, A. (2017) TD-DFT calculations of one- and two-photon absorption in Coumarin C153 and Prodan: attuning theory to experiment. Phys. Chem. Chem. Phys.: v. 19 i. 42 p. 28824-28833
- Mikhaylov, A., Lindquist, J., Callis, P., Kohler, B., Pahapill, J., de Reguardati, S., Rammo, M., Uudsemaa, M., Trummal, A., Rebane, A. (2017) Femtosecond two-photon absorption spectra and permanent electric dipole moment change of tryptophan, 2-aminopurine and related intrinsic and synthetic fluorophores. Proceedings SPIE: v. 10069
- Drobijev, M., Callis, P., Nifosi, R., Wicks, G., Stoltzfus, C. R., Barnett, L., Hughes, T. E., Sullivan , P., Rebane, A. (2015) Long- and short-range electrostatic fields in GFP mutants: Implications for spectral tuning. Scientific Reports: p. 13223
- Woods, L., Callis, P., Walker, R. (2015) Adsorption and Aggregation at Silica/Methanol Interfaces: The Role of Solute Structure. J. Phys.Chem C
- Xu, J., Chen, B., Callis, P. R., Muiño, P. L., Rozeboom, H., Broos, J., Toptygin, D., Brand, L., Knutson, J. R. (2015) Picosecond Fluorescence Dynamics of Tryptophan and 5‑Fluorotryptophan in Monellin: Slow Water−Protein Relaxation. J. Phys. Chem. B
- Callis, P. (2015) Simulating Electrostatic Effects on Electronic Transitions in Proteins. special issue. Molecular Simulation: v. 41 i. Special Issue p. 190–204
- Callis, P. (2014) Binding Phenomena and Fluorescence Quenching. II: Photophysics of Aromatic Residues and Dependence of Fluorescence Spectra on Protein Conformation. J. Mol. Struct. : v. 1077 p. 22–29
- Callis, P. (2014) Descriptive Quantum Principles of Fluorescence Quenching using a Supermolecule Jablonski Approach. special issue. Journal of Molecular Structure: v. 1077 i. Special issue p. 14-21
- Biesso, A., Xu, J. H., Muino, P., Callis, P., Knutson, J. (2014) Charge Invariant Protein-Water Relaxation in GB1 via Ultrafast Tryptophan Fluorescence,. Journal of the American Chemical Society: v. 136 i. 7 p. 2739−2747
Quantitative prediction of fluorescence quantum yields in proteins: Tryptophan
Tryptophan fluorescence intensity is widely used to monitor almost any imaginable change in protein structure. Tryptophan fluorescence intensity is widely used to monitor almost any imaginable change in protein structure. Using hybrid quantum mechanics-molecular mechanics (QM-MM) simulations we have recently shown that the full 30-fold range of Trp fluorescence quantum yields (and lifetimes) observed in proteins is due primarily to different rates of electron transfer from the excited indole ring to one of two nearest backbone amides. This heretofore puzzling dependence on protein environment arises mainly from the average local electric potential difference between the Trp ring and acceptor amide and from the amplitude of potential difference fluctuation caused by protein and solvent motions.
- We continue to pursue stronger documentation of the precise nature of the charge transfer state on the amide through higher level quantum calculations.
- We continue to apply our method to test and predict Trp fluorescence in a growing array of proteins.
- We are refining similar predictions for quenching caused by other protein residues, including protonated histidine, disulfide, amide side chains, and cysteine.
- We are studying the nature of the electron transfer quantum mechanical matrix element, which is a major factor in the quenching process.
Selected Publications:
Callis PR, Liu T:
Quantitative predictions of fluorescence quantum yields for tryptophan in proteins
J. Phys. Chem. B 108 4248-4259 (2004)
Xu J, Toptygin D, Graver KJ, Albertini RA, Savtchenko RS, Meadow ND, Roseman S, Callis
PR, Brand L, Knutson JR :
Ultrafast Fluorescence Dynamics of Tryptophan in theProteins Monellin and IIAGlc
J. Am. Chem. Soc. 128 ASAP (2006)
Callis PR, Vivian JT:
Understanding the variable fluorescence quantum yield of tryptophan in proteins using
QM-MM simulations. Quenching by charge transfer to the peptide backbone
Chem. Phys. Letters 369 409-414 (2003)+A49
Keywords:
Biophysical, Protein Chemistry
Quantitative prediction of fluorescence quantum yields in proteins: Flavins and dyes
Oxidized flavin cofactors exhibit an even wider range of fluorescence intensities and lifetimes than tryptophan. The source of this quenching is better understood: flavin quenching is almost always by electron transfer from nearby unexcited tryptophan and tyrosine to the excited state of the flavin. The same appears to be the case for many of the fluorescent dyes that are attached to proteins for various imaging and diagnostic purposes. We are modeling the efficiency of this process with the same tools we use for modeling Trp fluorescence.
Selected Publications:
Callis PR, Liu T :
Short Range Photoinduced Electron Transfer in Proteins: QM-MM Simulations of Tryptophan
and Flavin Fluorescence Quenching in Proteins
Chemi Phys. (2006) in press+A60+A25
Keywords:
Biophysical, Protein Chemistry
Quantitative prediction of fluorescence spectra and spectral relaxation of tryptophan in proteins.
The tryptophan fluorescence spectrum in proteins depends sensitively on the local environment covering the range 308-355 nm. We have previously shown that the wavelength can be predicted from QM-MM simulations. One must sum over all charged atoms of the protein and solvent to find the electric potential different from the 5- to the 6-membered ring of the Trp. Recently, there is considerable interesting in how rapidly the solvent and protein environment responds to the suddent large dipole increase caused by excitation of Trp. We are modelling the dynamics of this shift with our QM-MM simulations to see why controversial, long relaxation times appear in some experiments.
Selected Publications:
Xu J, Toptygin D, Graver KJ, Albertini RA, Savtchenko RS, Meadow ND, Roseman S, Callis
PR, Brand L, Knutson JR:
Ultrafast Fluorescence Dynamics of Tryptophan in theProteins Monellin and IIAGlc
J. Am. Chem. Soc. 128 ASAP (2006)
Vivian JT, Callis PR:
Mechanisms of Tryptophan Fluorescence Shifts in Proteins
Biophys. J. 80 2093-2109 (2001)
Keywords:
Biophysical
